Sensing and Monitoring
Molecular and cellular sensing for monitoring and diagnostic purposes
Fractionation of whole blood using a combination of dielectrophoresis and pinched co-flow for liquid biopsy
Doctoral researcher: Alexandra Rolland
Supervision: Christophe Vieu / Hervé Aubert (MINC, LAAS)
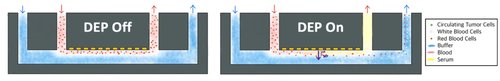
At all stages of Cancer disease, it has been shown that the tumor emits circulating tumoral cells (CTCs), exosomes and circulating tumoral DNA (ctDNA), into the bloodstream, which constitute as many biomarkers revealing the biology of the tumor. Liquid biopsy, the sampling and analysis of a biofluid, offers a minimally invasive way to analyze these markers, but isolating them reliably is challenging.
We here propose a device for the fractionation of a whole blood sample using a combination of dielectrophoresis and pinched co-flow. More specifically, this device resuspends all cells contained in the blood in a selected buffer enabling better filtration for CTC isolation using micro-filtration devices. In parallel, blood serum containing non cellular biomarkers such as ctDNA and exosomes can be recovered for downstream analysis. This device aims to enable the extraction of multiple cancer biomarkers from a single blood sample.
Collaboration: SmartCatch (Aline Cerf, Zacchari Ben Meriem), LAAS CNRS / équipe MINC (Hervé Aubert)
Large-Scale 3D Transcriptomic Mapping of Tumors
Post-doctoral researcher: Simon Dumas
Spatial transcriptomics has emerged as a powerful method to map gene expression within tissues, offering valuable insights into complex multicellular processes. However, current methods are restricted to 2D tissue sections. This project aims to develop a next-generation 3D spatial analysis method using DNA-barcoded microfabricated devices. By associating mRNA with spatial barcodes and decoding them through DNA sequencing, this approach will create large-scale 3D molecular maps, enabling the analysis of entire biological systems, such as tumors, without the need for tissue sectioning. First applied to pancreatic tumors — known for their spatial heterogeneity at different scales — the method seeks to enhance our understanding of 3D tissue architecture and interactions.
Collaborations: TBI (Emmanuelle Trévisiol) , GeT-Biopuces
Fundings: TIRIS Junior Fellowship Program 2023
Atto-Traces: Detection of molecular traces for Spatial and Environmental sciences using a superhydrophobic fluidic concentrator coupled with metallic plasmonic nano-antennas for Surface Enhanced Raman Spectroscopy analysis, in the femto-molar range.
Doctoral researcher: Sofien Ramos
Supervisor: Christophe Vieu / Emmanuelle Trevisiol (TBI)
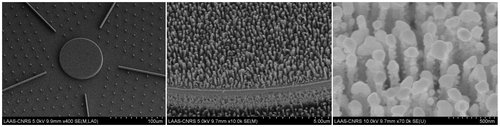
Nanofabricated device. Left side: view of the Super-hydrophobic surface with the guiding lines and the central analytical pedestal, middle image: zoom on the central pedestal equipped with Silicon nano-pillars, Right side: Silver plasmonic nanoparticles decorating the silicon nano-pillars for SERS analysis.
Text and Picture: Efficient superhydrophobic fluidic concentrators are developed using a hierarchical superhydrophobic surface on which the evaporation of a sessile droplet of water drives all the non-volatile elements it contains on a pre-defined micrometric analytical surface (pedestal of 80 µm). This hierarchical silicon surface exhibits a surface texture made of etched nano-pillars and consists of micro-pillars and guiding lines, arranged in a radial symmetry around the central pedestal. The guiding lines ensure the overall convergence of the sessile droplet towards the central pedestal during evaporation, while the nano-pillar texturing delays the Cassie-Baxter to Wenzel regime transition, until the edge of the droplet reaches the periphery of the pedestal. The full nanofabrication process also includes a thin film deposition of a noble metal such as Ag or Au which generates plasmonic nano-antennas on top of the silicon nano-pillars.This process has been optimized to generate the largest surface density of hot spots combined with high electromagnetic enhancement factors. The very high concentration capacity of the super-hydrophobic device combined with Surface Enhanced Raman Spectroscopy (SERS) analysis and a dedicated algorithm of spectra analysis enables the detection of various analytes diluted at concentrations down to the atto-molar range (10-18 mol/L). These devices are optimized and used for different types of application:
- For Spatial sciences and exobiology investigations, traces of life (DNA, amino acids, proteins, lipids) are detected by SERS and a space compatible instrumentation is developed.
- For Environmental sciences, water pollutants are analyzed in rough water samples with un-precedented limits of detection.
Collaborations: TBI, Toulouse Biotechnologies Institute, IRAP, Institut de Recherche en Astrophysique et Planétologie de Toulouse.
Publications:
V. Fabre, F. Carcenac, A. Laborde, J.B. Doucet, P. Louarn, C. Vieu and E. Trévisiol, Hierarchical Superhydrophobic Device to Concentrate and Precisely Localize Water-Soluble Analytes: A Route to Environmental Analysis. Langmuir 38, 14249–14260 (2022) https://hal.inrae.fr/hal-03880333
Funding: INSA-Toulouse, Emerging projects call
Automated analytical method for monitoring micro-organisms in marine waters
Doctoral researcher: Eva Agranier (MICA, LAAS)
Supervisor: Vincent Raimbault (MICA, LAAS) / Julia Baudart (OBS)/ Bastien Venzac
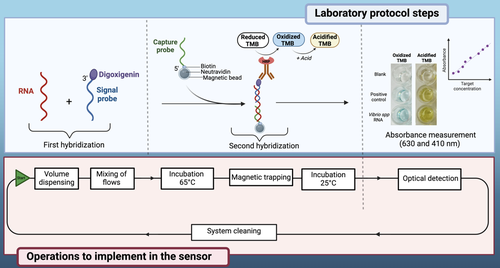
Aquatic environments play a fundamental role in sustaining life on Earth and contribute to the global economy through fishing, aquaculture, and coastal tourism, while also serving as a major source of food for the world’s population. Nevertheless, the presence of pathogenic microorganisms in coastal waters can negatively impact human and animal health, as well as marine ecosystems. Bacteria of the Vibrio spp genus, for example, are among the many species pathogenic to humans and marine fauna. While their seasonal abundance dynamics are now recognized, a better understanding on a finer temporal scale, as well as their spatial distribution, requires the use of in situ measuring instruments for real-time monitoring of these bacterial populations in water. In this context, we propose the development of an innovative miniaturized measuring tool for in situ monitoring of microbial populations, based on a molecular method for genetic identification of bacterial taxa, which was previously developed in the laboratory. This new measuring tool is designed to be used by non-specialized personnel and will be capable of autonomous biomonitoring in all types of aquatic environments, thus supporting enhanced biomonitoring of oceans and seas.
Collaborations :
Observatoire Océanologique de Banyuls sur mer – Sorbonne Université - CNRS : Carmem-Lara Manes, Renaud Vuillemin, Michel Groc
Publications:
Da-Silva E, Barthelmebs L, Baudart J. Development of a PCR-free DNA-based assay for the specific detection of Vibrio species in environmental samples by targeting the 16S rRNA. Environ Sci Pollut Res. 2017;24:5690–700.
Lace A, Byrne A, Bluett S, Malaquin L, Raimbault V, Courson R, et al. Ion chromatograph with three‐dimensional printed absorbance detector for indirect ultraviolet absorbance detection of phosphate in effluent and natural waters. J Sep Sci. 2022;45(5):1042-50
Fundings: Groupement de Recherche Océan et MERs (GDR OMER) – Projet MORTIMER
AFM-FluidFM-ML for single cell transcriptomic
Doctoral researcher: Koutayba Saada
Supervisor: Etienne Dague, Laurent Malaquin