Structural Bioinformatics
Development of algorithms for modeling and designing flexible biomolecules such as proteins
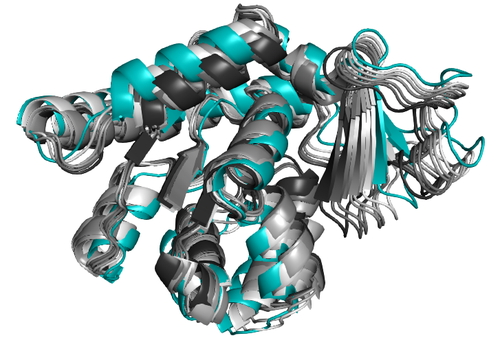
Biomolecules serve as the fundamental building blocks of life, with proteins standing as versatile workhorses among them. Proteins play pivotal roles in various biological processes, acting as catalysts, structural components, and signaling agents within cells. Many of these functional roles relay on their flexibility.
The diverse functionalities of proteins have led to their extensive use in various biotechnological applications. In fields like medicine, proteins serve as targets for drug development. Additionally, in the realm of biopharmaceuticals, proteins such as antibodies are harnessed for therapeutic and diagnostic purposes. The catalytic properties of proteins are widely exploited in industrial biotechnologies, enabling more efficient and environmentally friendly production processes. In all these domains, the possibility to optimize the properties of natural proteins or to design new proteins de novo paves the way for groundbreaking advancements in science and technology.
The study and design of proteins requires a synergistic combination of experimental and computational methods. While experimental techniques like X-ray crystallography, NMR spectroscopy, and cryo-electron microscopy offer crucial insights into protein structures and functions, they often provide snapshots that may not capture the full dynamic range of protein behavior. Computational methods complement these experimental techniques by elucidating the dynamic behavior and conformational flexibility of proteins.
In this context, we carry out interdisciplinary research, in collaboration with biologists, biophysicists, chemists, physicists and mathematicians, and develop new methods for modeling and designing proteins (as well as other molecules), with an emphasis on new approaches to deal with their flexibility.
These methods are made available to the scientific community via web servers (https://moma.laas.fr/), or in the form of binaries and source code (https://gitlab.laas.fr/moma).